
 Ph.D. Student in AIDD
Ph.D. Student in AIDDI am a Ph.D. candidate at Sichuan University, specializing in deep geometric learning models for biomolecules and drug design. My research integrates knowledge- and data-driven approaches to accelerate drug development and advance solutions for combating diseases.
Warning
Problem: The current name of your GitHub Pages repository ("Solution: Please consider renaming the repository to "
http://".
However, if the current repository name is intended, you can ignore this message by removing "{% include widgets/debug_repo_name.html %}" in index.html.
Action required
Problem: The current root path of this site is "baseurl ("_config.yml.
Solution: Please set the
baseurl in _config.yml to "Education
-
 SiChuan UniversityDepartment of Medical Chemistry, West China School of Pharmacy
SiChuan UniversityDepartment of Medical Chemistry, West China School of Pharmacy
Ph.D. student in AIDDSep. 2023 - present -
 SiChuan UniversityDepartment of Medical Chemistry, West China School of Pharmacy
SiChuan UniversityDepartment of Medical Chemistry, West China School of Pharmacy
M.S. student in Drug DesignSep. 2019 - Jul. 2023 -
 SiChuan UniversityWest China School of Pharmacy
SiChuan UniversityWest China School of Pharmacy
B.S. student in PharmacySep. 2016 - Jul. 2019
Honors & Awards
-
Doctoral Fellowship in Youth Science and Technology Talent Program, China Association for Science and Technology, 2026-20272026
-
Student membership, China Association for Artificial Intelligence (CAAI), 2026-20272026
-
Sichuan University National Scholarship for Doctoral Students, 2024-20252025
-
Best Poster 1st class, Asia Hub for e-Drug Discovery 2025 (AHeDD2025)2025
-
Certificate of Presentation for Virtual Graduate Students Symposium in Asia-Pacific Region on Computational Chemistry, ACS Spring 20252025
-
Outstanding Presentation Award at the Third National Pharmaceutical Graduate Academic Symposium2023
-
Second Prize at the Ninth Sichuan University Internet+ College Student Innovation and Entrepreneurship Competition2023
Selected Publications (view all )
Pharmacophore-Oriented 3D Molecular Generation towards Efficient Feature-Customized Drug Discovery.
Peng, J.*, Yu, J.-L.*, Yang, Z.-B.*, Chen, Y.-T.*, Li, G.-B.# (* equal contribution, # corresponding author)
Nature Computational Science 2025
PhoreGen, a novel pharmacophore-oriented 3D molecular generation method, uses asynchronous updates and message-passing to integrate ligand-pharmacophore mapping, producing chemically reasonable, diverse, and drug-like molecules with high binding affinity. It successfully identified new bicyclic boronate inhibitors for metallo- and serine-β-lactamases and first-in-class covalent inhibitors for metallo-nicotinamidases, demonstrating its potential for feature-customized drug discovery.
Pharmacophore-Oriented 3D Molecular Generation towards Efficient Feature-Customized Drug Discovery.
Peng, J.*, Yu, J.-L.*, Yang, Z.-B.*, Chen, Y.-T.*, Li, G.-B.# (* equal contribution, # corresponding author)
Nature Computational Science 2025
PhoreGen, a novel pharmacophore-oriented 3D molecular generation method, uses asynchronous updates and message-passing to integrate ligand-pharmacophore mapping, producing chemically reasonable, diverse, and drug-like molecules with high binding affinity. It successfully identified new bicyclic boronate inhibitors for metallo- and serine-β-lactamases and first-in-class covalent inhibitors for metallo-nicotinamidases, demonstrating its potential for feature-customized drug discovery.
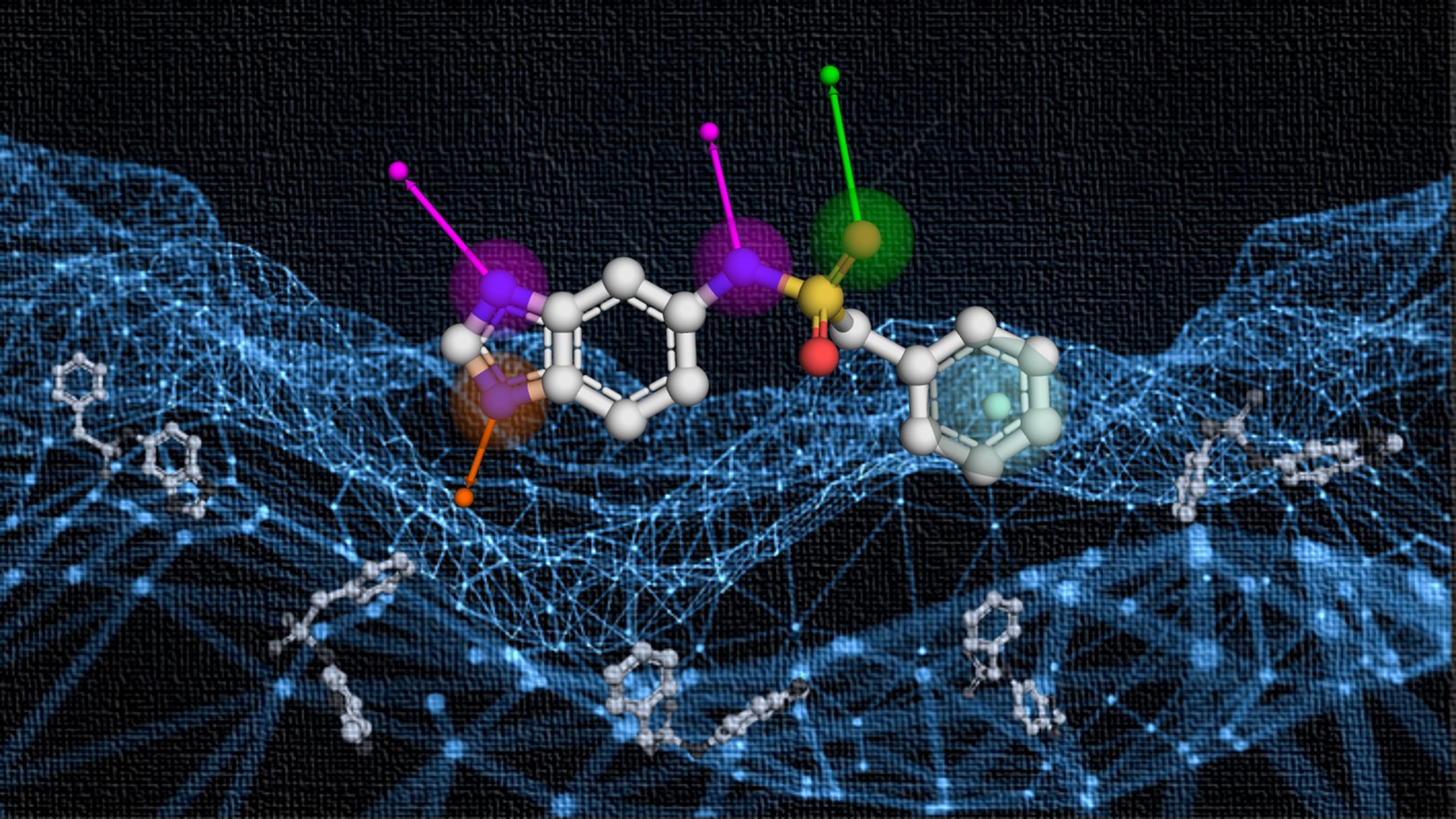
Knowledge-Guided Diffusion Model for 3D Ligand-Pharmacophore Mapping.
Yu, J.-L., Zhou, C., Li, G.-B.# (# corresponding author)
Nature Communications 2025
A knowledge-guided diffusion framework for ‘on-the-fly’ 3D ligand-pharmacophore mapping, named DiffPhore, which achieves state-of-the-art performance in predicting ligand binding conformations, surpassing traditional pharmacophore tools and several advanced docking methods.
Knowledge-Guided Diffusion Model for 3D Ligand-Pharmacophore Mapping.
Yu, J.-L., Zhou, C., Li, G.-B.# (# corresponding author)
Nature Communications 2025
A knowledge-guided diffusion framework for ‘on-the-fly’ 3D ligand-pharmacophore mapping, named DiffPhore, which achieves state-of-the-art performance in predicting ligand binding conformations, surpassing traditional pharmacophore tools and several advanced docking methods.
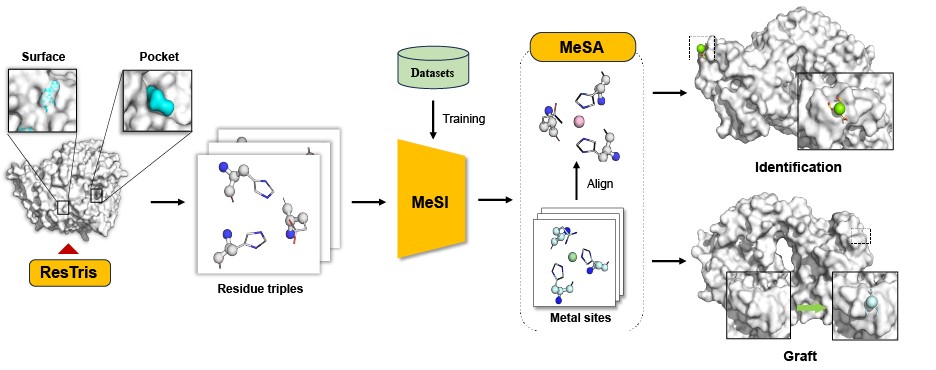
Geometric deep learning-enabled metal-binding site identification and grafting.
Yu, J.-L.*, Wang, Y.-G., Peng, J., Wu, J.-W., Zhou, C., Li, G.-B.# (* equal contribution, # corresponding author)
Fundamental Research 2024
MeSiteIG, a geometric deep learning tool, enables metal-binding site identification and grafting using E3-equivariant graph neural networks, achieving high accuracy and speed (~300 samples/second) in predicting metal-binding residues, identifying overlooked protein metal-binding sites, and designing novel metalloproteins by grafting metal sites onto antibodies and protein pockets.
Geometric deep learning-enabled metal-binding site identification and grafting.
Yu, J.-L.*, Wang, Y.-G., Peng, J., Wu, J.-W., Zhou, C., Li, G.-B.# (* equal contribution, # corresponding author)
Fundamental Research 2024
MeSiteIG, a geometric deep learning tool, enables metal-binding site identification and grafting using E3-equivariant graph neural networks, achieving high accuracy and speed (~300 samples/second) in predicting metal-binding residues, identifying overlooked protein metal-binding sites, and designing novel metalloproteins by grafting metal sites onto antibodies and protein pockets.
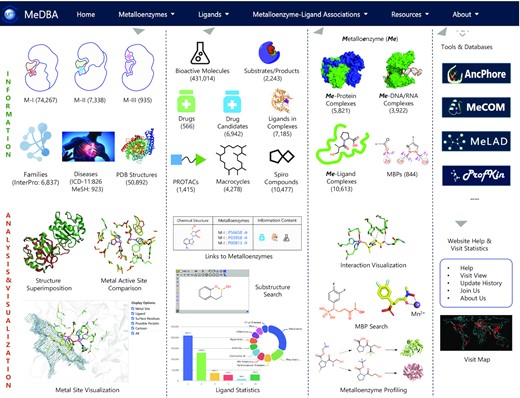
MeDBA: the Metalloenzyme Data Bank and Analysis Platform.
Yu, J.-L.*, Wu, S.*, Zhou, C., Dai, Q.-Q., Schofield, Christopher J., Li, G.-B.# (* equal contribution, # corresponding author)
Nucleic Acids Research 2023
This work has expanded the scope of metalloenzyme-specific knowledge and services, by forming a versatile platform, termed the Metalloenzyme Data Bank and Analysis (MeDBA), which provides comprehensive information on metaloenzyme activities, expression profiles, family and disease links.
MeDBA: the Metalloenzyme Data Bank and Analysis Platform.
Yu, J.-L.*, Wu, S.*, Zhou, C., Dai, Q.-Q., Schofield, Christopher J., Li, G.-B.# (* equal contribution, # corresponding author)
Nucleic Acids Research 2023
This work has expanded the scope of metalloenzyme-specific knowledge and services, by forming a versatile platform, termed the Metalloenzyme Data Bank and Analysis (MeDBA), which provides comprehensive information on metaloenzyme activities, expression profiles, family and disease links.
Deep learning in target prediction and drug repositioning: recent advances and challenges.
Yu, J.-L.*, Dai, Q.-Q.*, Li, G.-B.# (* equal contribution, # corresponding author)
Drug Discovery Today 2022
This review details the advancements and applications of deep learning in innovative drug discovery, covering protein structure prediction, drug target prediction, drug-target interaction prediction, drug synthesis route design, de novo drug design, and ADMET prediction, while summarizing current challenges and potential solutions to guide future development.
Deep learning in target prediction and drug repositioning: recent advances and challenges.
Yu, J.-L.*, Dai, Q.-Q.*, Li, G.-B.# (* equal contribution, # corresponding author)
Drug Discovery Today 2022
This review details the advancements and applications of deep learning in innovative drug discovery, covering protein structure prediction, drug target prediction, drug-target interaction prediction, drug synthesis route design, de novo drug design, and ADMET prediction, while summarizing current challenges and potential solutions to guide future development.